WP1
WP4
The main focus of work package four is to study different components of the immune response in relation to their location within the tissue using deep learning algorithms. A convolutional neural network will be developed to detect and quantify multiple structures in histopathological slides of kidney biopsies showing various types of immunological rejection. This algorithm will be made robust for staining variations between different centers.
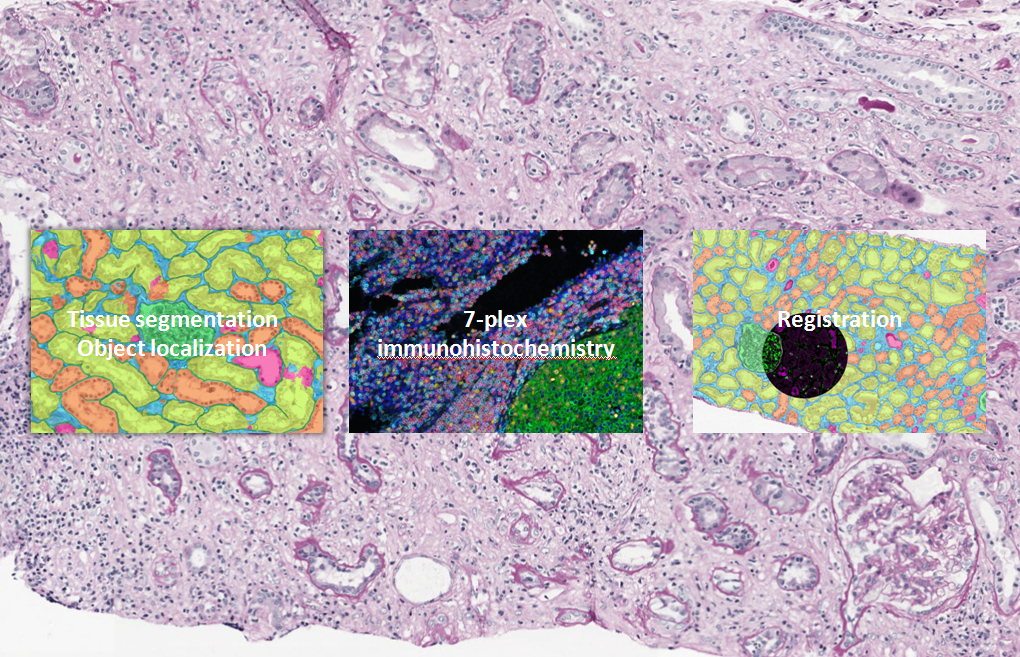
Moreover, a multiplex panel composing of 6 different markers will be designed and validated to investigate the microenvironment during rejection, with a main focus on cellular rejection and (i-)IFTA. In order to be able to separate the 6 markers, the multiplex stained slides will be scanned using a multi-spectral camera. We will create an additional algorithm for the detection of immune cells in the resulting multispectral images. The information retrieved from mIHC shall be automatically transferred to a fully automatically segmented consecutive slide which will provide additional spatial information of the inflammatory components in relation to their environment.
WP5 – Knowledge-based image analysis for multiplexed IHC in the kidney-specific microenvironment
The main objectives of WP5 are to integrate immunological expert knowledge into an image-specific medical ontology and generate novel image analysis algorithms for histopathological images. The specific image analysis methods developed will be built in a modular way, in order to be integrated into a workflow. This knowledge-driven image analysis workflow will be developed and histopathology image-related ontologies generated, based on methods and experiences adapted from geosciences.
To be as relevant as possible, novel tools have to strongly rely on expert knowledge, which is in our previous experience the key to offer useful and accurate automatic image analyses tailored to a specific scientific question. An important challenge is to consequently model this knowledge and to use automatic interpretation tools complementing rather than replacing conventional expert slide evaluation. From a technological point of view, the goal of this work package is to address the following questions: (1) How does the pathologist interpret microscopic images and how can we model the knowledge he uses? (2) What information can we automatically extract from the images, and where are currently limitations of digital slide analysis? (3) How can we match and integrate the information extracted from the images and the expert knowledge?
The workpackage is divided into three tasks:
- Task 5.1: Ontology instantiation and implementation for AAM in renal transplantation (rTx)
- Task 5.2: Knowledge-based image analysis of macrophages in the microenvironment of the kidney

- Task 5.3: Contribution to clinical validation
WP6
WP6 interacts with all partner institutions and supports the joint goals by sample retrieval, image data generation, and clinical integration of novel image analysis workflows.
The primary objective of WP6 is to push the limits of current immune cell detection in kidney biopsies through experimental biomarker discovery in collaboration with WP3 and by innovative digital pathology tools and workflows that integrate formalized expert knowledge in interaction with WP5 and WP4. The activities support analysis of the underlying dynamic processes that is performed in WP1 and WP2.
WP6 will translate highly multiplexed exploratory AAM marker assays using fluorescence microscopy and multispectral image analysis into innovative biomarker workflows feasible in translational research.
Our tasks include :
- Task 6.1 Multiplexed IHC assays to detect Mɸ/T-cell subpopulations in their renal tissue context
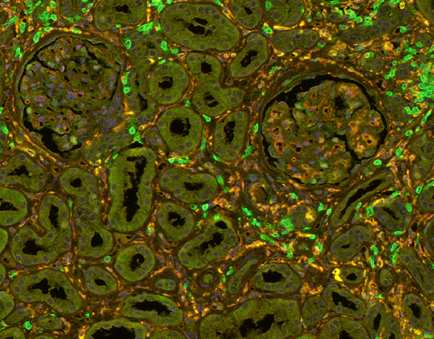
Fig 1: T-cells (green and Macrophages (yellow) in a transplanted kidney
- Task 6.2 Repository of enriched cell populations (Mɸ and T-cells) by microdissection of paraffin-embedded renal tissues
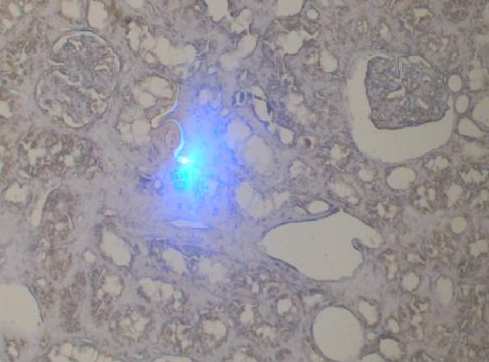
Fig 2: Laser-microdissection of fibrotic regions in a transplanted kidney
- Task 6.3 Alignment and harmonization of analysis workflows and data management between two clinical centers (MHH and RUMC)
